NeDRexApp: Use cases
In the following, we demonstrate the utility of NeDRex in five different use cases employing available functionalities in NeDRexApp. You can find more information for each use case in the NeDRex publication.
For all use cases except for the HD use case with BiCoN, a network containing Gene-Disorder and Disorder-Disorder
association types needs to be imported from NeDRexDB first.
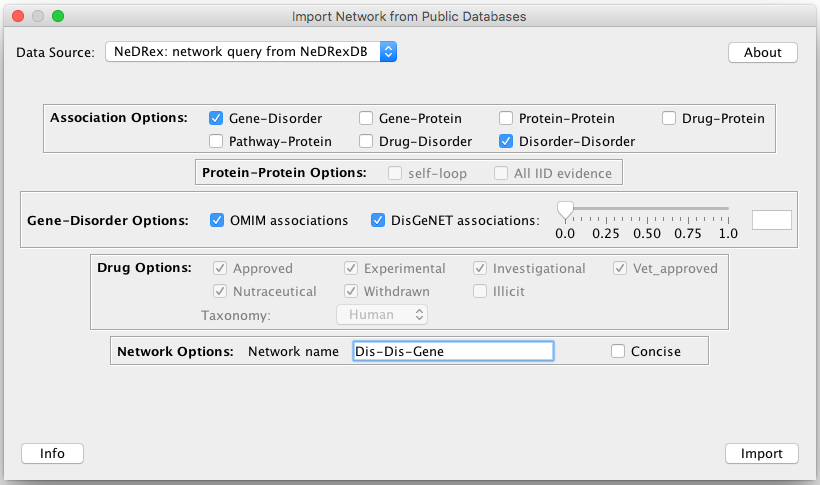
You can get this network from NeDRexDB by clicking on
File –> Import –> Network from Public Databases... –> NeDRex: network query from NeDRexDB and selecting
the associations (see Import Network from Public Databases for an explanation of the parameters):
Additional information about the different methods (MuST, DIAMOnD, BiCoN, Closeness Centrality, TrustRank) can be found in the NeDRex: Algorithms section.
Please note that the results can slightly vary due to updates in the underlying databases.
Identifying disease pathways for OC: MuST
In this use case we show how to extract biologically meaningful pathways for a disease under study. We use the ovarian cancer (OC) associated genes from NeDRexDB and construct the corresponding disease module using the MuST algorithm.
Using the Quick Select function (Apps –> NeDRex), we can easily find and select the node corresponding
to ovarian cancer. Select the network you imported and run the Quick Select function, choose Disorder as node type
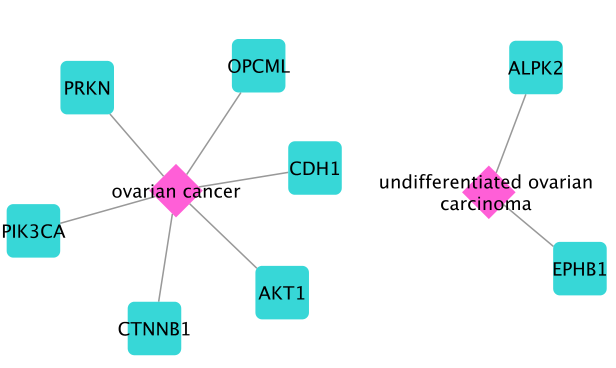
and type in ovarian ca. Like this, you can find the term ovarian cancer and undifferentiated ovarian carcinoma.
Add both terms to the list and click on Select in Network. The nodes with the mondo IDs mondo.0008170 and mondo.0006477
should now be selected.
In order to obtain the disease genes, execute the function Get Disease Genes. For this use case, leave the box for selecting subtypes unchecked.
The result contains a subnetwork with OC disease nodes and their associated genes AKT1, ALPK2, CDH1, CTNNB1, EPHB1, OPCML, PIK3CA and PRKN.
Next, we will construct the disease module using the MuST algorithm with these seed genes. First, we need to select all
genes with Select Nodes –> All nodes of specific type –> Gene. Then, we run MuST
(Disease Module Identification –> Run MuST) with the following parameters setting: number of Steiner trees = 5,
maximum number of iterations = 5, no hub penalty.
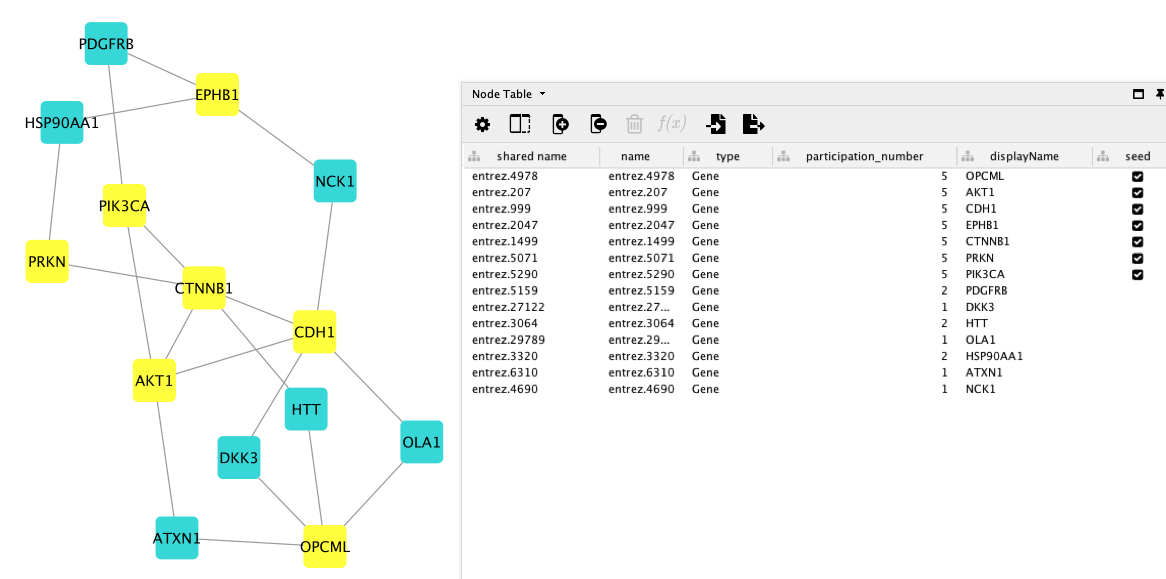
The obtained disease module contains newly identified connector genes (ATXN1, HTT, HSP90AA1, PDGFRB, NCK1, OLA1 and DKK3) which, together with the seed nodes, participate in relevant ovarian cancer pathways that could not be retrieved with the seed genes only.
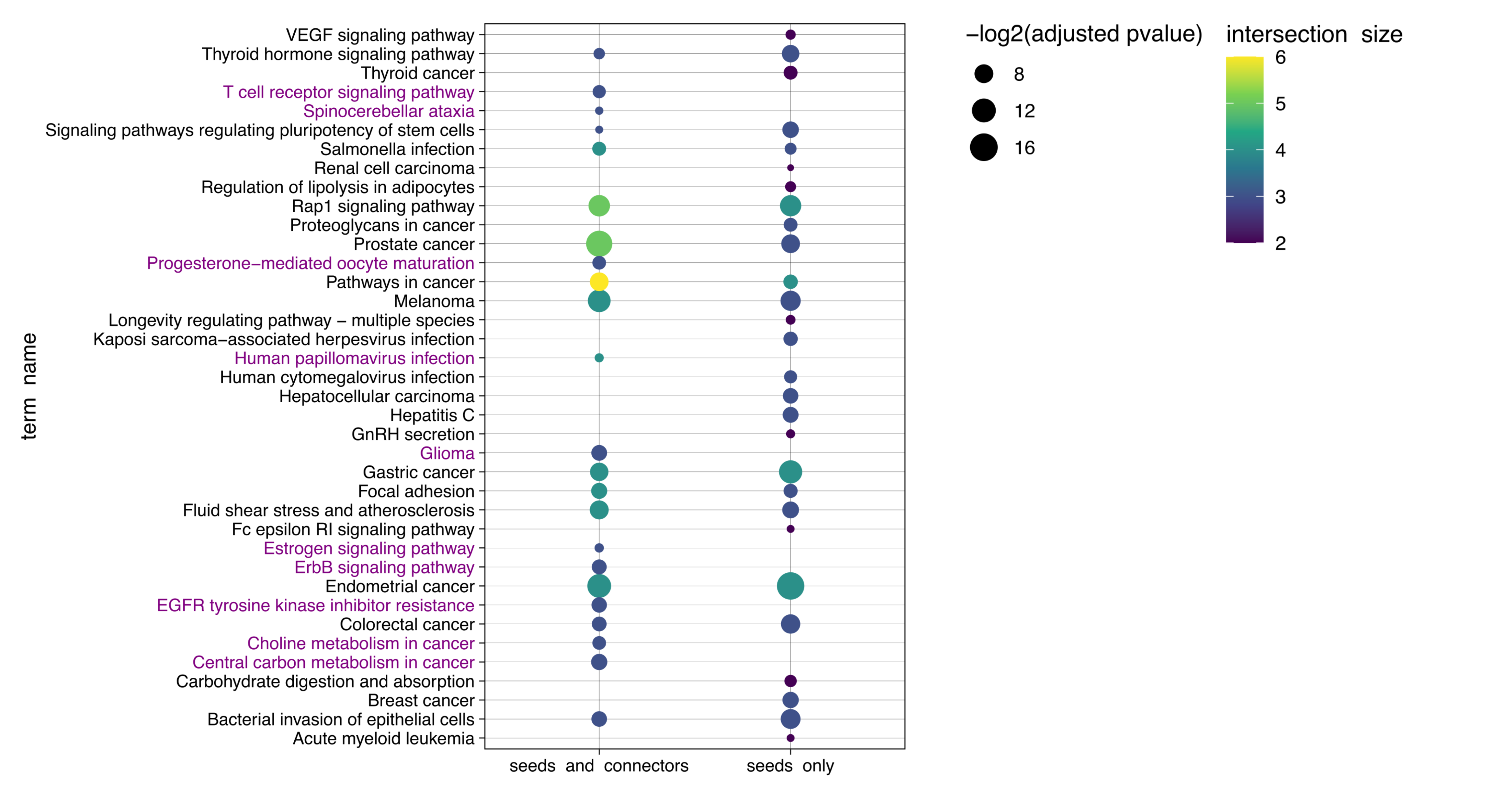
Taking a closer look at the identified genes, we can see that in particular, ovary-specific, hormone-related and cancer pathways are found. Using the g:Profiler enrichment tool and the KEGG pathway as data source, we find that the OC module is enriched in the “progesterone-mediated oocyte maturation” and the “Estrogen signaling pathway”, which are both involved in oocyte maturation.
The following figure illustrates the comparison of KEGG enriched pathways obtained with seed and connector genes vs. obtained with only seed genes. Pathways which could only be retrieved after adding connector genes are marked in purple.
Furthermore, the examination of the connector genes identified by MuST reveals that the PDGFRB gene has been proposed as a therapeutic target in OC (ref).
The results in this use case show that, using MuST, we were able to identify a disease module containing genes associated with meaningful biological pathways (ovary-specific and cancer-associated pathways) for OC.
Identifying therapeutic drugs for IBD: MuST + Closeness Centrality
In this use case we demonstrate how to recover known and potential therapeutic drugs for a disease under study. We start by selecting
inflammatory bowel disease (IBD) with ID mondo.0005265 via the Quick Select function (node type Disorder, type in the disease name,
Add to the list and Select in Network).
Next, we obtain the disease genes with the Get Disease Genes function. For this use case, leave the box for selecting subtypes unchecked. The following 30 associated genes are obtained for IBD:
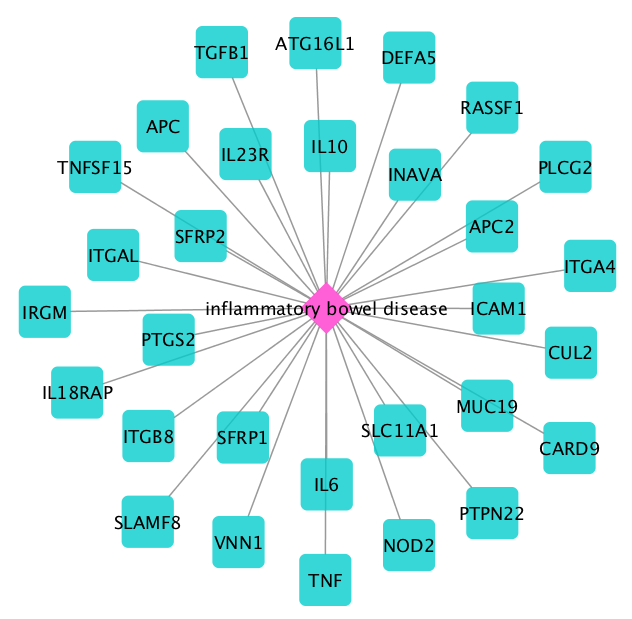
We first select all disease genes as seeds (Select Nodes –> All nodes of specific type –> Gene).
Then, we run MuST
(Disease Module Identification –> Run MuST) with the following parameters setting: number of Steiner trees = 5,
maximum number of iterations = 5, no hub penalty.
The obtained disease module has 87 genes:
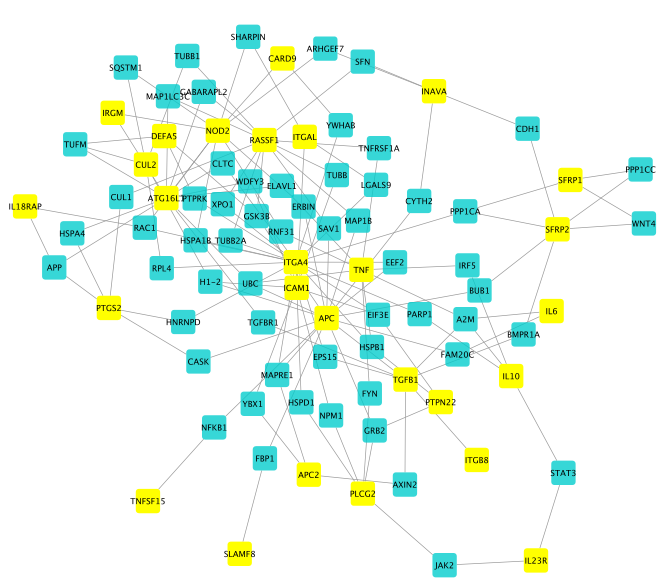
We now prioritize drug candidates targeting the disease module by using one of the drug ranking functionalities, here Closeness centrality.
We first select all genes in the module as seeds and run Drug Prioritization –> Rank drugs with Closeness centrality
with the following parameters: include only direct and approved drugs, result size = 50.
The obtained IBD disease module derived by MuST method, combined with its targeting 25 top-ranked drugs by closeness centrality:
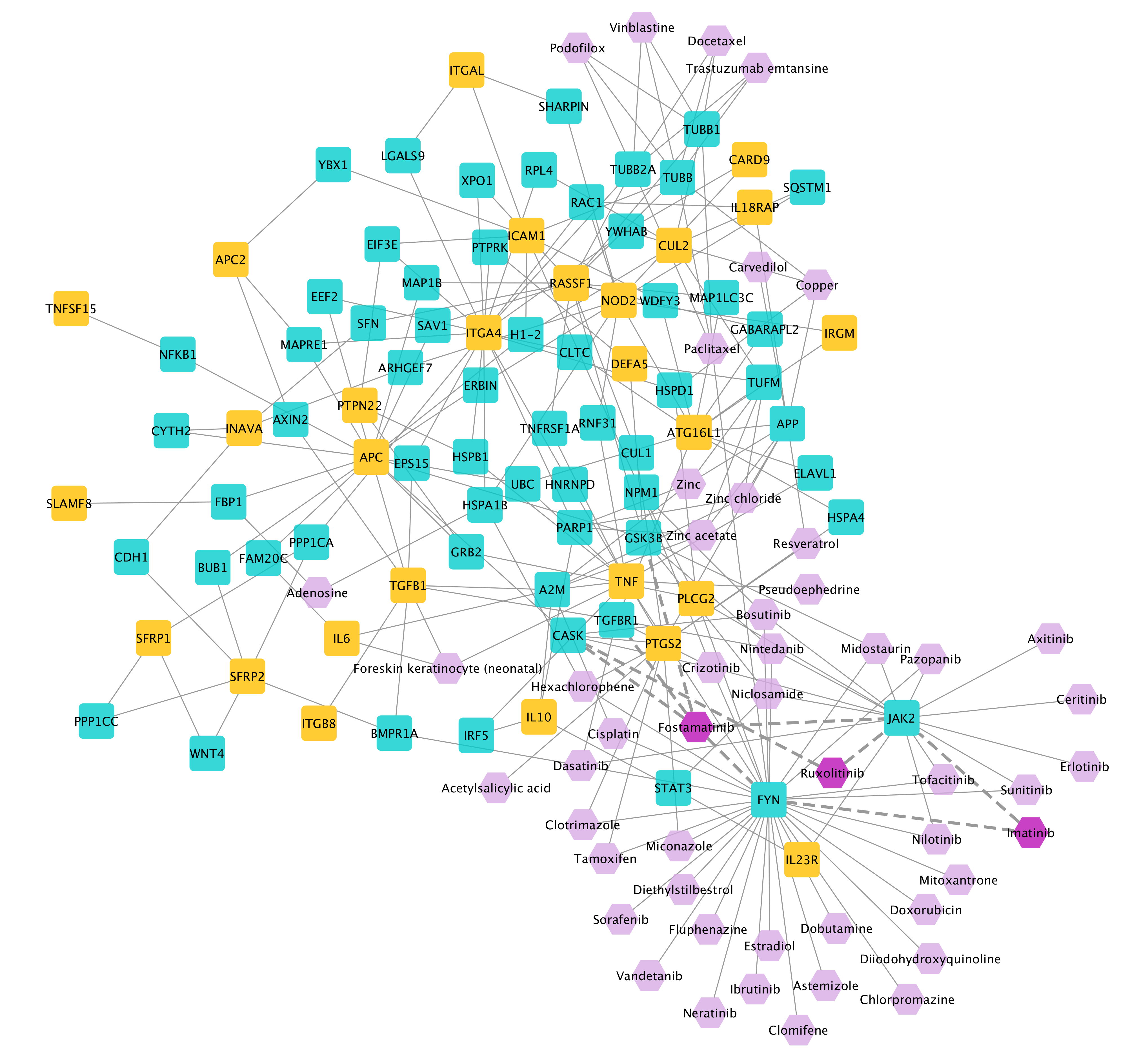
Three drugs among the top-ranked drugs, namely, Fostamatinib, Ruxolitinib, and Imatinib are identified, which have also been indicated for the treatment of IBD or whose relevance to IBD is supported by literature evidence.
The results presented in this use case provide further motivation to explore the potential of other top-ranking drugs in the treatment of IBD derived by using NeDRexApp.
You can also find a screencast video showing you step-by-step how to reproduce the IBD use case here:
Drug target and drug identification for PE: DIAMOnD + TrustRank
In this use case we demonstrate how we can uncover drug targets for pulmonary embolism (PE) disease using the DIAMOnD algorithm and subsequently recover drugs indicated for treatment of PE.
First, we retrieve all genes associated to PE by first selecting the disease in the network (Quick Select function,
node type Disorder, search for pulmonary embolism (disease) or mondo.0005279, Add to the list and Select in Network) and then
executing the function Get Disease Genes without checking the subtype box.
The following 12 genes associated to PE are obtained:
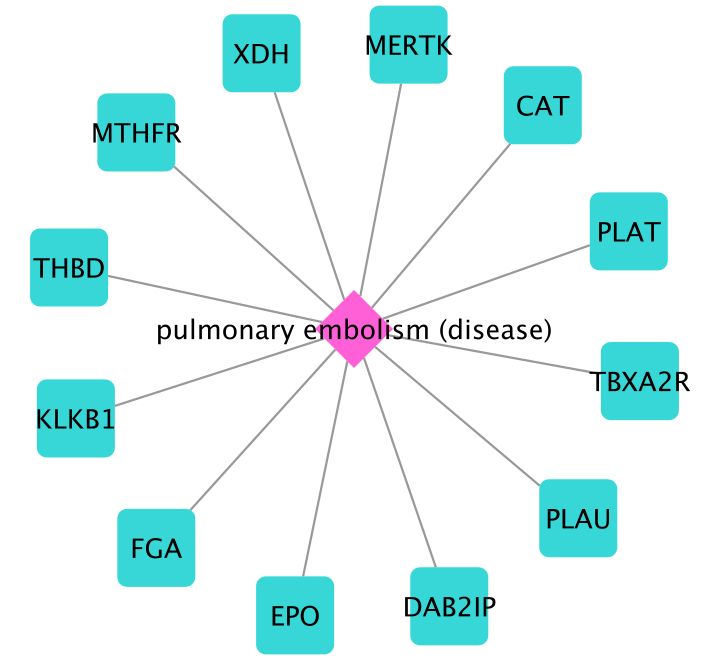
After selecting all obtained genes as seeds by Select Nodes –> All nodes of specific type –> Gene, we now run DIAMOnD (Disease Module Identification –> Run DIAMOnD)
to identify the disease module for PE with the following parameters: number of iteration = 20, weight of seeds = 1, return all edges in the result disease module = True.
The DIAMOnD algorithm returns a subnetwork of 32 genes representing the underlying mechanistic pathways for PE:
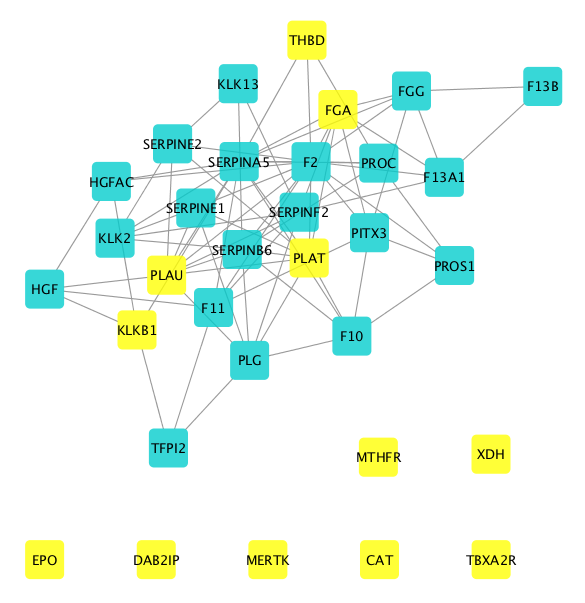
If you are interested to get an overview and find all the drugs targeting this module, you need to first select all nodes in the module and then
run Drug Prioritization –> All drugs targeting the selection. You will see in the result network there are 283 drugs targeting this module. Therefore, it’s recommended
to proceed with the ranking step.
For this specific use case, we are interested in whether we can find relevant drugs even without including the initial seed genes, i.e.
just selecting the DIAMOnD nodes. Therefore, you need to go to the network corresponding to the disease module and only select the nodes that
are not labeled as seeds from the node table (e.g. sort by seed attribute, mark all non-seed rows, right click and choose Select nodes from selected rows).
Then run TrustRank via Drug Prioritization –> Rank drugs with TrustRank with the following parameters:
include only direct and approved drugs, damping factor = 0.85, result size = 100.
The PE disease module (excluding the initial seeds) derived by DIAMOnD, combined with its targeting 50 top-ranked drugs:
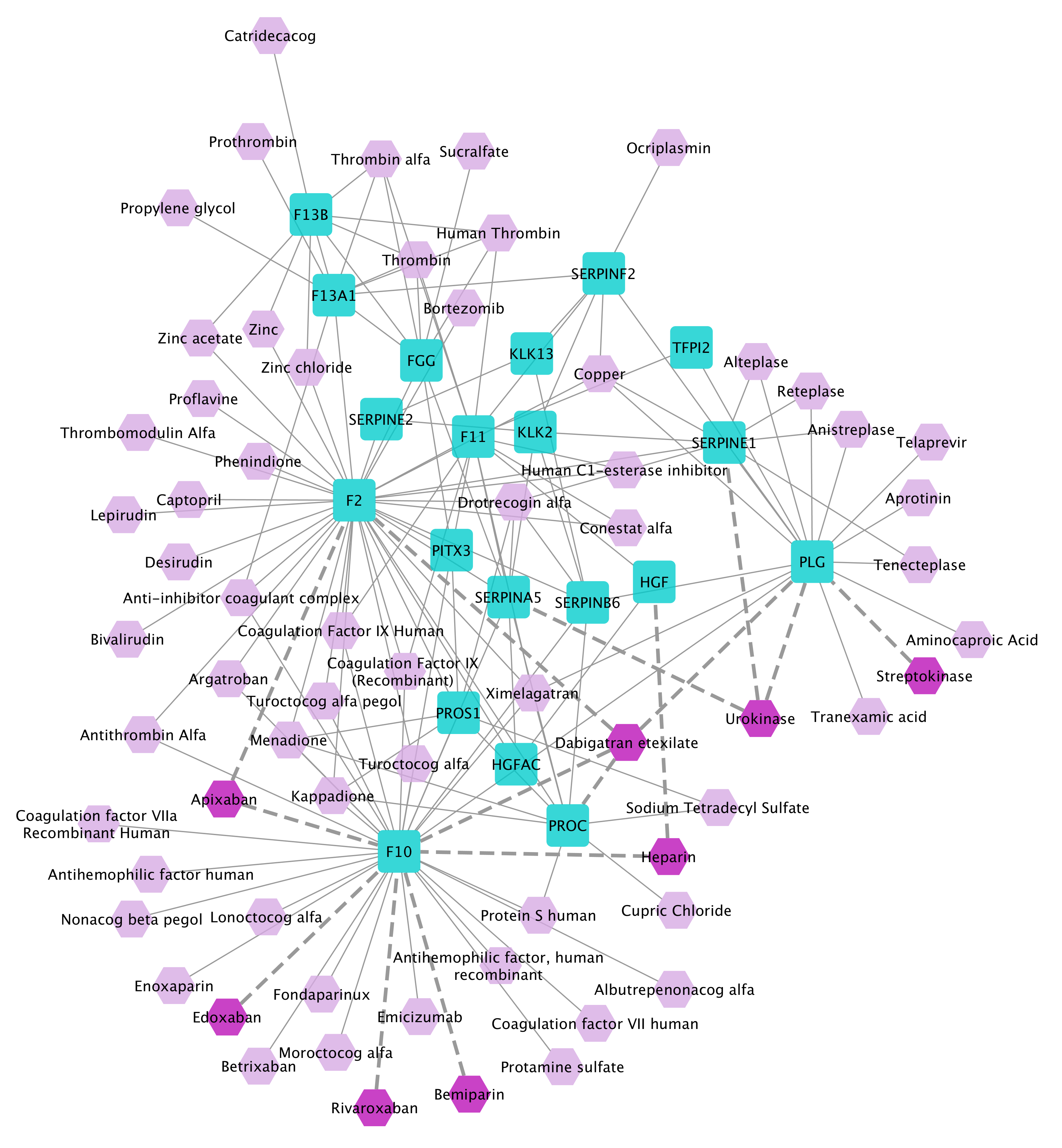
We find Bemiparin, Edoxaban, Apixaban, Dabigatran etexilate, Heparin, Rivaroxaban, Streptokinase, and Urokinase among the 50 top-ranked drugs (colored dark purple). All of these drugs are indicated to reduce the risk of stroke and systemic embolism and are known to be used to treat PE. Furthermore, five drugs registered in ClinicalTrials.org for evaluation in treatment of PE, namely, Alteplase, Enoxaparin, Fondaparinux, Tenecteplase, and Tranexamic acid are found at the top of the ranked list.
F10 (a key gene in the coagulation cascade) which is not among the initial set of PE-associated genes and was returned in the disease module is identified as one of the drug targets for PE. Apixaban, Bemiparin and some other top-ranked drugs target this gene. Likewise, PLG (helping in dissolving the fibrin of blood clots) was not among the initial seeds and identified in the returned disease module as drug target for PE.
With this use case we firstly demonstrated that the NeDRexApp can extract disease-related mechanistic pathways, which can contain possible targets for candidate drugs. Secondly, drugs which in practice are prescribed for treatment of PE or are under evaluation in clinical trials are among the top-ranked drugs obtained by the drug ranking algorithms.
You can also find a screencast video showing you step-by-step how to reproduce the PE use case here:
Disease module and drug identification for HD: BiCoN + TrustRank
In this use case we demonstrate how we can identify disease module for Huntington’s Disease (HD) using the BiCoN algorithm and subsequently identify potential repurposable drugs for this disease.
For BiCoN we do not need to import a network from NeDRexDB, instead it’s required to supply gene expression data in the format explained in Run BiCoN.
You should first download the HD expression file for this use case (Huntington’s disease gene expression data from GEO,
accession number GSE3790, patients with Vonsattel grades 2–4 and healthy controls).
Then run BiCoN via Disease module identification function –> Run BiCoN using the downloaded expression file as input and
with the following parameters: minimal solution subnetwork size = 10, maximal solution subnetwork size = 15.
BiCoN returns a connected subnetwork of genes which is the union of two identified gene clusters (red and blue). The resulting subnetwork is
colored with respect to its difference in expression patterns in patients clusters. You can also find the patients grouping table and the biclustering heatmap in the Results Panel:
After selecting all genes from BiCoN module as seeds by Select Nodes –> All nodes of specific type –> Gene, we can find drugs targeting
this module by running TrustRank via Drug Prioritization –> Rank drugs with TrustRank with the following parameters:
include only direct and approved drugs, damping factor = 0.85, result size = 50.
The HD disease module derived by BiCoN, together with its targeting 50 top-ranked drugs.
Among the 50 top-ranked drugs we find three drugs that are prescribed to alleviate the symptoms of Huntington’s Disease, namely Thorazine (Chlorpromazine), Memantine, and Lamotrigine. There are also some other drugs in this list that have a strong connection to Huntington’s Disease such as Donepezil, Decamethonium, Betahistine, Fluoxetine, and Pitolisant (colored dark purple).
Hypothesis-driven drug repurposing for AD
In this use case, we show how NeDRex can be used to extract possibly repurposable drugs which are indicated for diseases that are known to be associated with the disease of interest. More specifically, using Alzheimer’s disease (AD) as an example, we show that we can retrieve new potential treatments with an original indication for hypertension, diabetes mellitus and hyperlipidemia.
For this use case, we import a network containing Gene-Disorder and Disorder-Disorder associations with a
stricter DisGeNet cutoff score of 0.5:
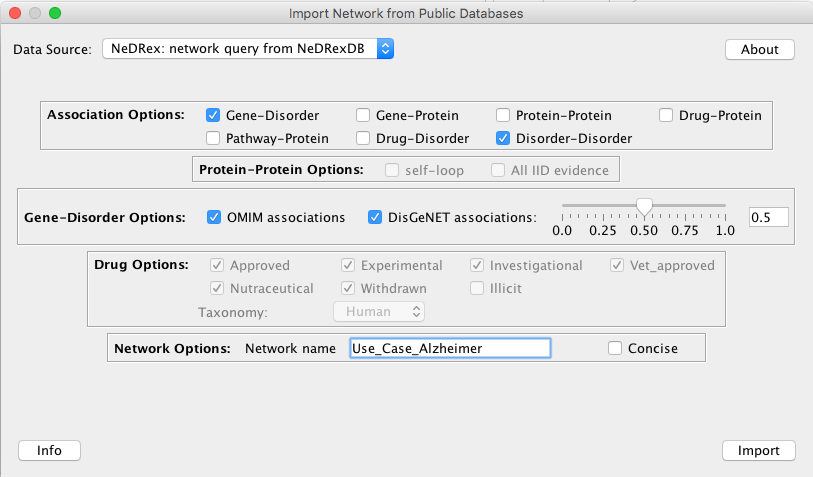
Please notice that visualizing and applying a layout to the network, built based on the parameters above, take a bit long (~5 mins). This needs to be done only once.
Hypertension as original indication
Here, we demonstrate how NeDRexApp can identify repurposable drugs directly from the genes associated for the new indication as a starting point. (Get drugs targeting the disease genes)
With the Quick Select function (node type Disorder), we now look for Alzheimer disease with ID mondo.0004975, add it and select it in
the network.
Since “Alzheimer disease” is an umbrella term for many diseases in MONDO, we check the box Include all subtypes of disorders when
executing Get Disease Genes.
In this way, 39 disease genes are found:
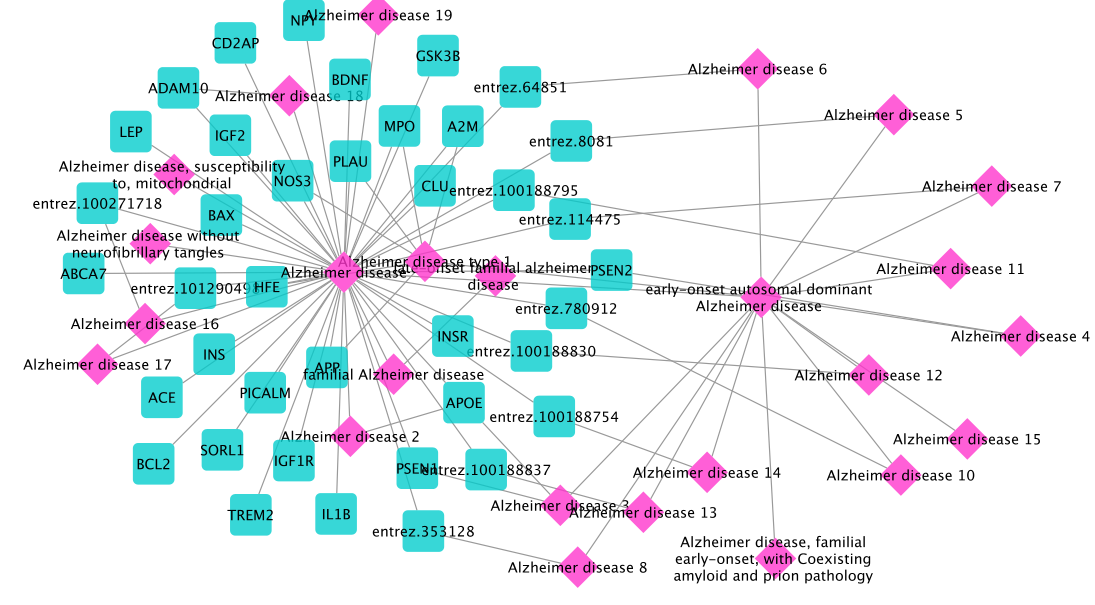
We directly rank the drugs targeting the returned disease genes without finding the disease module first as we did for previous use cases. For this,
first select all genes by Select Nodes –> All nodes of specific type –> Gene and then run Drug Prioritization –>
Rank drugs with Closeness Centrality with the following parameters: include only direct and approved drugs, result size = 100.
In the result network, you can see the drugs with their ranks in the node table. Interestingly, in this network we can find Telmisartan (ranked 26th) which is a known angiotensin II receptor blocker (ARB) originally indicated to treat high blood pressure and has been tested in clinical trials to assess its efficacy for the treatment of AD. Studies show that drugs used to treat hypertension, including ARBs, decrease the risk and slow the progression of AD.
This use case shows that it is possible with NeDRexApp to retrieve potentially repurposable drugs directly from the associated genes of the new indication.
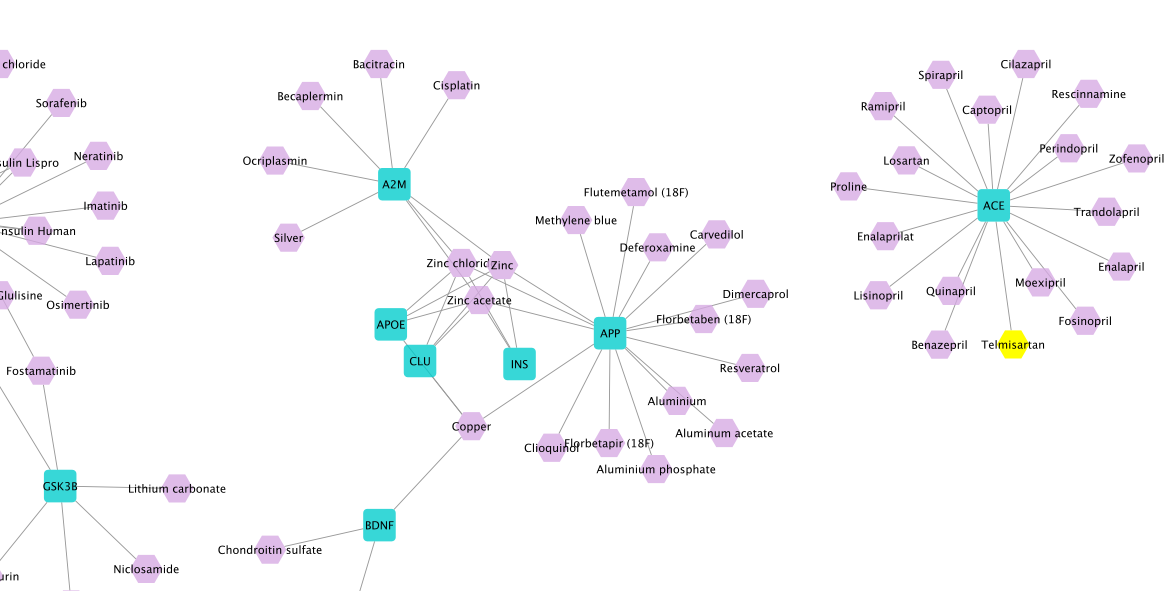
Diabetes as original indication
Medications indicated for diabetes mellitus are potential treatments of Alzheimer’s disease since the glucose metabolism plays a key role in neural function. Several drugs have been tested in vitro, in vivo and in clinical trials, namely, insulin (INS), Insulin Detemir, and Insulin Glulisine (insulin analogues). These drugs produce their effect by interacting with the insulin receptor (INSR) and are considered as disease modifying drugs.
In this use case, we show that our platform is capable of retrieving this shared molecular mechanism and these drugs.
We retrieve all AD associated genes as we did in the above use case. 39 genes are returned. Similarly, we get the disease
genes for diabetes with the term diabetes mellitus (disease) (mondo.0005015). 88 genes are returned.
We now obtain the intersection of the two networks (two sets of disease genes) using Cytoscapes Merge function (Tools –> Merge –> Networks...):
add the two networks to Networks to Merge, select Intersection and click on Merge.
The intersection results in two genes: INS (entrez.3630) and INSR (entrez.3643). Then, we find all drugs targeting these two genes by selecting both nodes and running
Drug Prioritization –> All drugs targeting the selection. The resulting network contains 32 drugs:
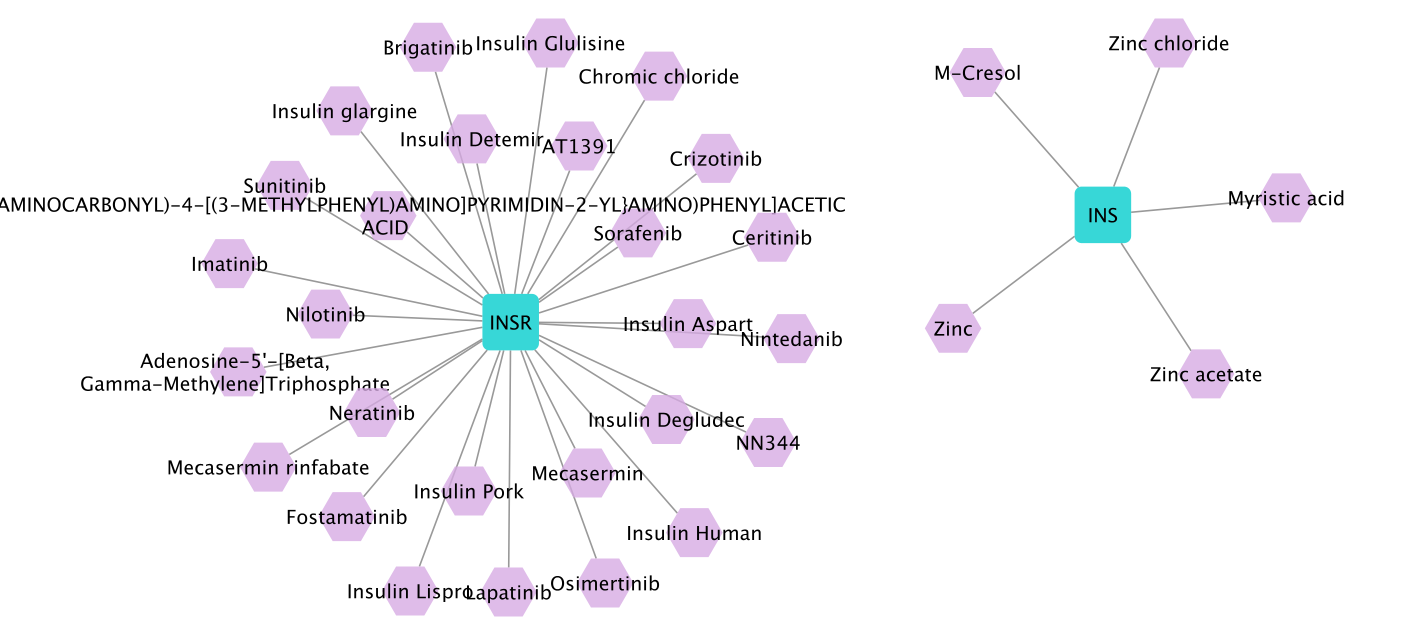
Among these drugs are Insulin, Insulin Detemir, Insulin Glulisine. Note that, in this use case, we did not use any network algorithms to extract the drug repurposing candidates but only leveraged the data integration functionalities provided by NeDRexApp to investigate the initial hypothesis of relation between AD and diabetes.
Hyperlipidemia as original indication
With this example, we show how to search for potentially repurposable drugs by retrieving drugs that indirectly target the intersection of disease modules for two diseases, namely, hyperlipidemia and Alzheimer’s disease (AD). We use the hyperlipidemia-associated genes, since the lipid and cholesterol metabolism has been linked with progression of Alzheimer’s disease.
First, we retrieve all hyperlipidemia-associated genes by selecting the disease in the graph using the Quick Select
function (node type: Disorder, term to add: hyperlipidemia (disease) or mondo.0021187) and then applying the Get Disease Genes function
(check subtype option box). The result contains 19 genes:
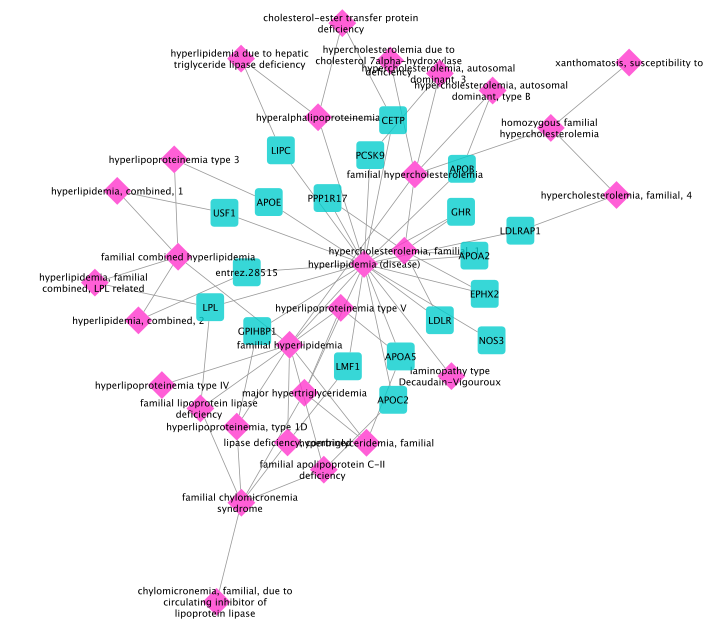
Next, we derive the disease module using DIAMOnD. For this, we first select all genes in the graph
(Select nodes –> All nodes of specific type –> Gene) and then run DIAMOnD via Disease Module Identification –> Run DIAMOnD
with the following parameters: number of iteration = 200, weight of seeds = 1, return all edges in the result disease module = False. This returns a network containing 218 genes.
Similarily, we derive the Alzheimer’s Disease module:
Quick Select function (node type:
Disorder, term to add:Alzheimer diseasewithmondo.0004975)Get Disease Genes (check subtype option box) -> returns 39 associated genes
Run DIAMOnD via
Disease Module Identification–>Run DIAMOnDwith the same parameters as we used for hyperlipidemia. This returns a network with 228 genes.
We are now interested in the intersection between these two modules. We obtain this by
using Cytoscapes Merge function (Tools –> Merge –> Networks...):
add the two networks to Networks to Merge, select Intersection, and click on Merge.
The intersection contains 7 genes: A2M, APOE, APP, CLU, IGF2, NOS3 and PLAU. Notably, all of them are Alzheimer’s disease genes and some are well-characterized drivers of this disease. Importantly, A2M, APP, CLU, IGF2 and PLAU are not among the hyperlipidemia associated genes, they are retrieved only after obtaining the disease module with DIAMOND. This demonstrates that in some cases, using only the disease associated genes is not enough to uncover the molecular mechanisms shared between diseases and using the disease module provides a more complete landscape of the disease.
Next, to retrieve the drugs directly targeting the overlapping genes (direct drugs) or their vicinity (indirect drugs),
we use closeness centrality with the option of including indirect drugs: We select all 7 nodes and then run
Drug Prioritization –> Rank drugs with Closeness Centrality with the parameters: include only direct drugs = False, include only approved drugs = True,
result size = 50.
The resulting network contains 76 drugs. We find Gemfibrozil among the top-ranked drugs (rank 6), which is originally indicated for the treatment of hyperlipidemia. Gemfibrozil is being tested in clinical trials and there are evidences of potential effectiveness of this drug for the treatment of AD. This drug does not directly target any of the 7 overlapping genes, and can be retrieved by using the indirect mode.
The following figure illustrates the AD and hyperlipidemia disease modules (top left and top right, respectively) derived by DIAMOnD using the corresponding disease genes (orange nodes). The intersection of the disease modules is shown in the middle. Gemfibrozil indirectly targets the intersection through the genes TTR, CYP2C8, and LPL (bottom). To allow better visualisation, subsets of actual networks corresponding to the disease modules are shown here:
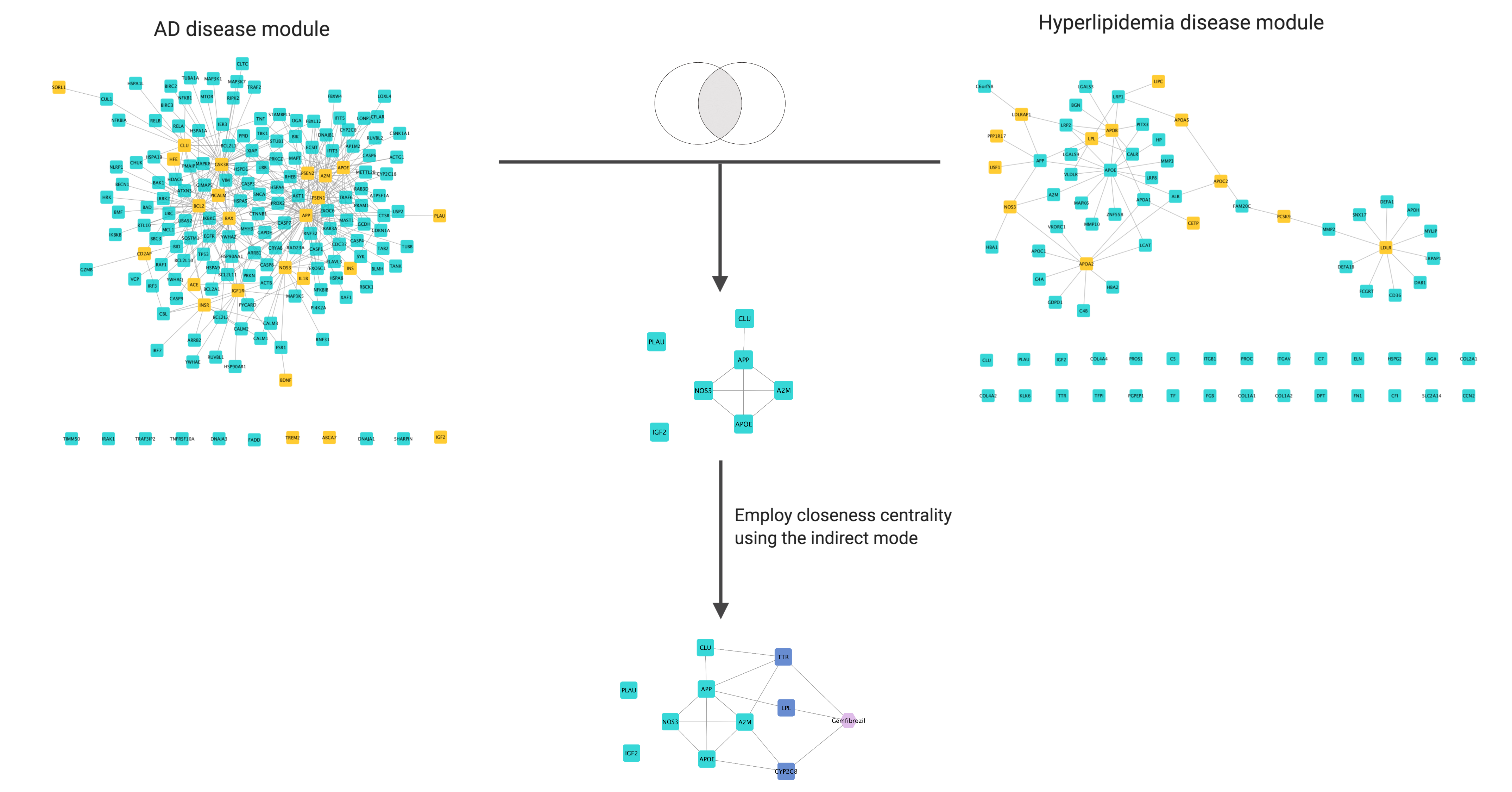
The indirect drugs can be interpreted as drugs targeting genes which are closely related to the seeds; in this case, Gemfibrozil targets TTR, CYP2C8 and LPL, which interact with APOE, A2M, CLU and APP, suggesting that this drug could produce the desired effect by affecting several targets which altogether affect the key genes of Alzheimer’s disease and hyperlipidemia.