Get ready to use the NeDRexApp
Use the NeDRexApp
Make sure you have Cytoscape installed. If not, please follow the instructions on their website. Please note that NeDRexApp is optimized for Cytoscape 3.
You can download NeDRexApp from the Cytoscape AppStore. Alternatively, you can install
the app by running the Cytoscape tool and finding NeDRex in the list of all apps via Apps –> App Manager.
Now, all the functions of the app should be available to you under Apps –> NeDRex.
Access the NeDRexDB via NeDRexApp
Using the NeDRexApp, networks from NeDRexDB can be imported using the Import Network from Public Databases function. The following nodes and edges from NeDRexDB are available via the app:
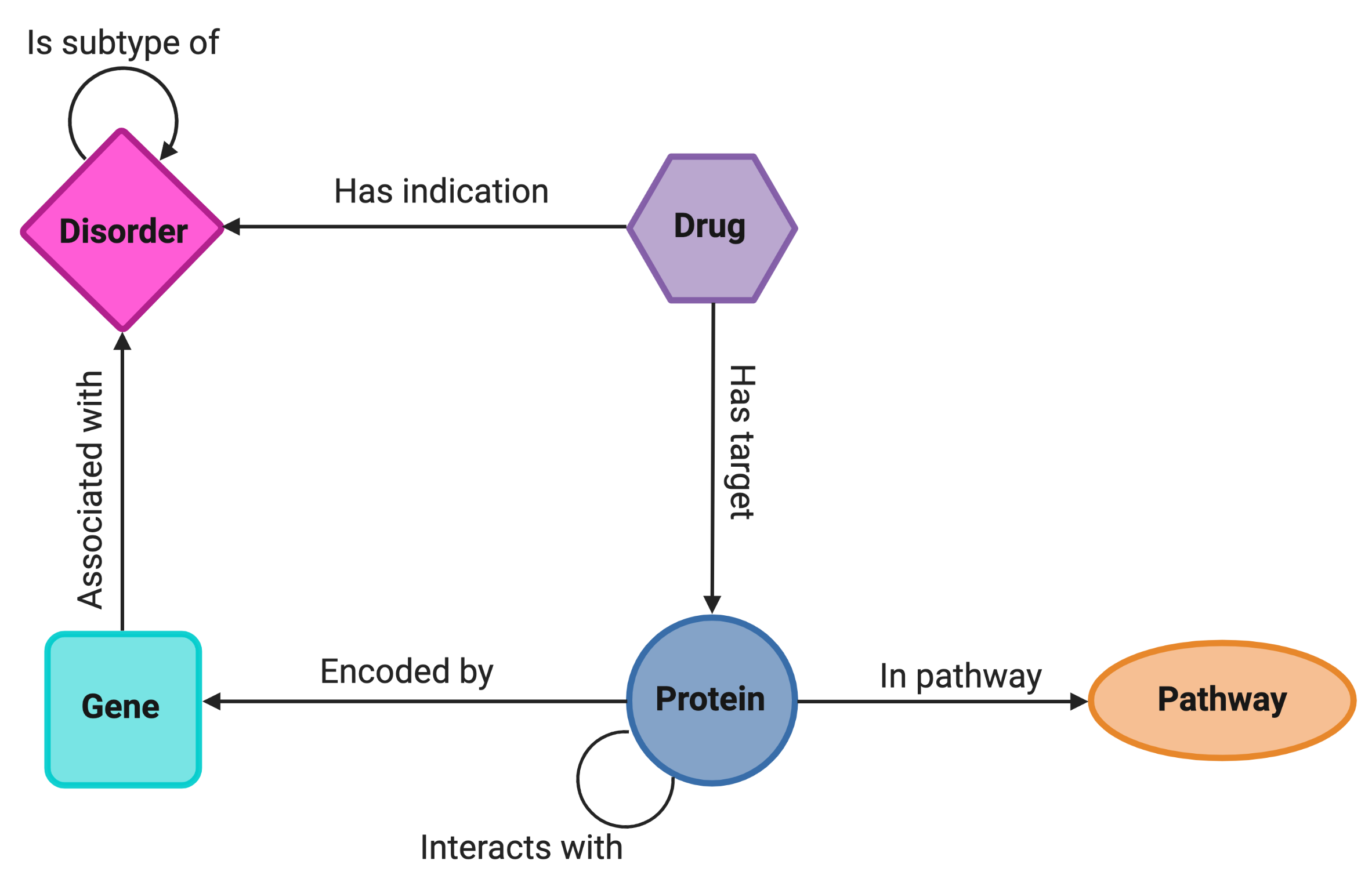
The NeDRexDB can be also be accessed in two ways without using the app: via NeDRexAPI or a Neo4j endpoint.
Import a network from NeDRexDB
You can import a heterogenous network constructed from NeDRexDB by going to File –> Import –> Network from Public Databases....
See the Import Network from Public Databases section for a thorough explanation of all parameters.
Choose NeDRex: network query from NeDRexDB as Data Source. You now have various options for customizing your network:
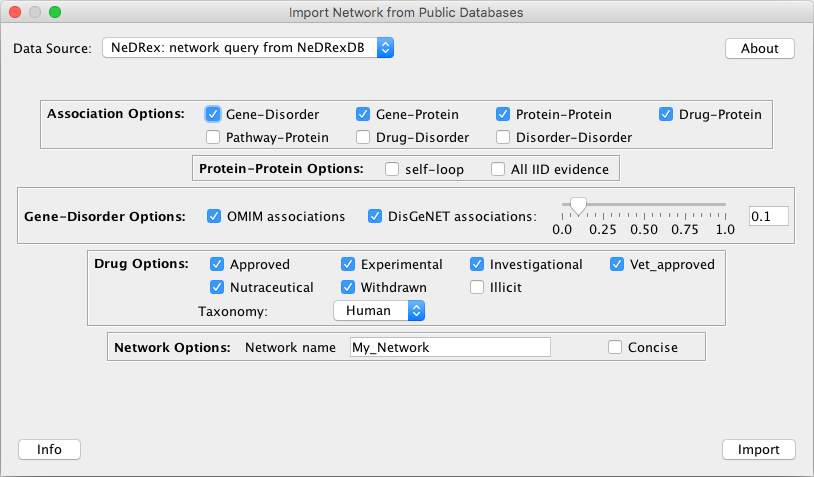
Import the network into Cytoscape by clicking on the Import button.
Load a network from file
If you do not want to use a network from NeDRexDB or you saved a customized NeDRexDB subnetwork locally, you can
load the network into Cytoscape via File –> Import –> Network from File. If you have saved your network as a Cytoscape session
file (cys format), you can load your network into Cytoscape via File –> Open.
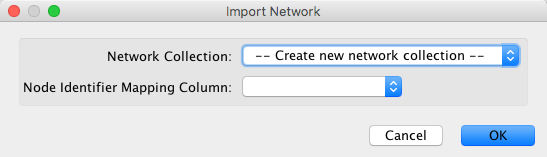
If you already have some other loaded networks in Cytoscape, when loading a new network, it is recommended to select
--Create new network collection-- option when prompted.
Please note that NeDRexApp is optimized for working with networks from NeDRexDB. If you would like to use externally created networks, make sure the format matches ours. The most important requirement is that for a heterogeneous network the type of nodes (such as Gene, Protein, Disorder, Drug, Pathway) needs to be specified as an attribute for nodes.